Lab to Table: Identifying the Genes Responsible For Muscle Tissue Development in Cows
April 13, 2021 - 4 min read
This article is about my recent project for the Ottawa Regional Science Fair. A more detailed look at the research methodology and findings can be found here.
What distinguishes a muscle cell from a fat cell or a skin cell? The answer is “gene expression.” More precisely, the particular combination of genes that are “turned on or off” in a cell which dictates cellular morphology and function. Although genome sequencing and analysis have been employed vastly in the study of disease and pharmaceuticals, there has been little application in the emerging field of cellular agriculture.
The process of cultivating meat requires exposing stem cells to the environmental cues necessary for them to proliferate and then differentiate into the specialized cells which eventually mature into the tissue characteristic of meat.
As most consumers are familiar with, the majority of meat products are composed of muscle tissue with small quantities of fat. A challenge then becomes finding a way to ensure these initial stem cells reliably differentiate into some combination of muscle cells and fat cells.
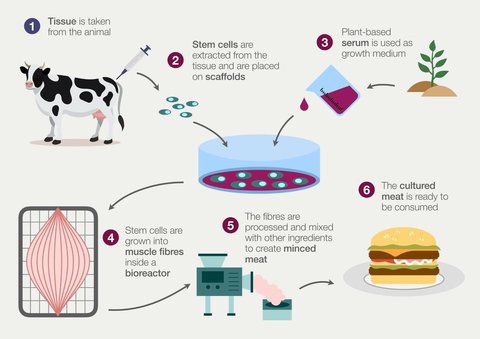
In thinking about how this can be done, the natural place to start is by considering how the process is already accomplished in vivo, as most in vitro bioprocesses are based on replicating the existing bodily functions responsible for the development of specialized tissue.
In living organisms, cell function operates according to the central dogma of molecular biology, which specifies that DNA transcribes mRNA which translates into proteins. These proteins are what dictate the role of that cell and is essentially what characterizes a cell as a blood cell as opposed to a neuron, for instance. The transcriptome of a cell is the set of genes which are translated from mRNA into proteins, and so while almost every cell in the mammalian body has the same genome, they may have different “isoforms” of a transcriptome. This ultimately leads to different sets of proteins being created, thus leading to distinct cell specializations.
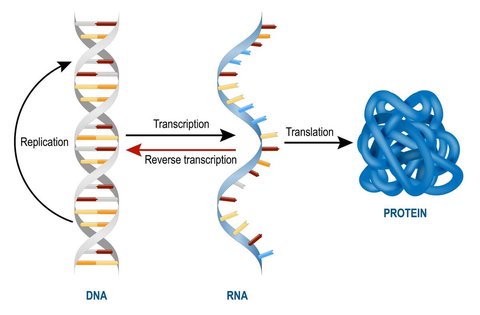
As such, a change in a cell’s function is caused by a change in its transcriptome as a response to external or internal stimuli such as development or exposure to a drug. During this process, specific genes are either up-regulated (expressed more) or down-regulated (repressed more). A gene is considered significantly differentially expressed if it is upregulated or downregulated beyond a predetermined threshold. Differential gene expression analysis is the process of identifying such genes by comparing two or more transcriptomes.
This year, I entered my project Lab to Table: A differential gene expression analysis of RNA-sequenced bovine stem cells & myocytes into the Ottawa Regional Science Fair. The purpose of this study was to design a bioinformatics pipeline to conduct a differential gene expression analysis on the transcriptomes of a bovine embryonic stem cell (ESC) and an adult myocyte (precursor to muscle tissue) in order to identify the genes responsible for bovine myocyte differentiation. The idea is that if we can find what these genes are, we can selectively activate them within stem cells so that in a cellular agriculture bioprocess they reliably differentiate into myocytes characteristic of meat that consumers would be willing to buy.
The two samples used here were obtained using RNA-sequencing (similar to genome sequencing but for mRNA) and are available on the NCBI Genbank database. They both came from bovine longissimus muscle — one from an ESC and the other from a myocyte. The differential gene expression analysis was run using a pipeline (series of algorithms) on the Galaxy Project public web server pictured below.
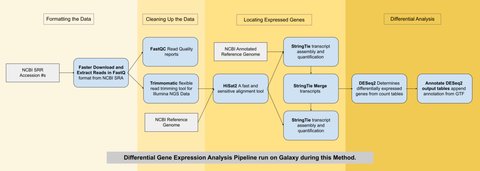
Running this pipeline and filtering the data resulted in 23 genes that were significantly differentially expressed: 10 of which were upregulated and 13 of which were downregulated over the process of development from a bovine ESC into a myocyte. These genes were then queried against the DAVID database which contains the functional annotation of genes across many organisms. Nine genes in particular stood out as being related to the development of muscle tissue.
In terms of next steps for this particular project, it would be useful to actually selectively upregulate or downregulate a subset of these genes within an ESC and monitor how the cell’s differentiation into a myocyte is impacted in practice. It would also be interesting to run a similar study regarding differentiation into adipocytes to further explore the possibility of creating cultivated products which have a balance of both tissue types. Comparing the gene analogs of different species of cows or the transcriptomes of cows raised on different diets would help to understand how the resulting modifications in gene expression influence organoleptic properties such as taste and texture of the resulting meat.


















